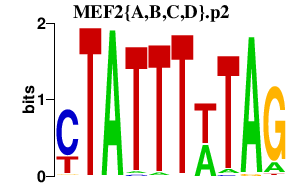
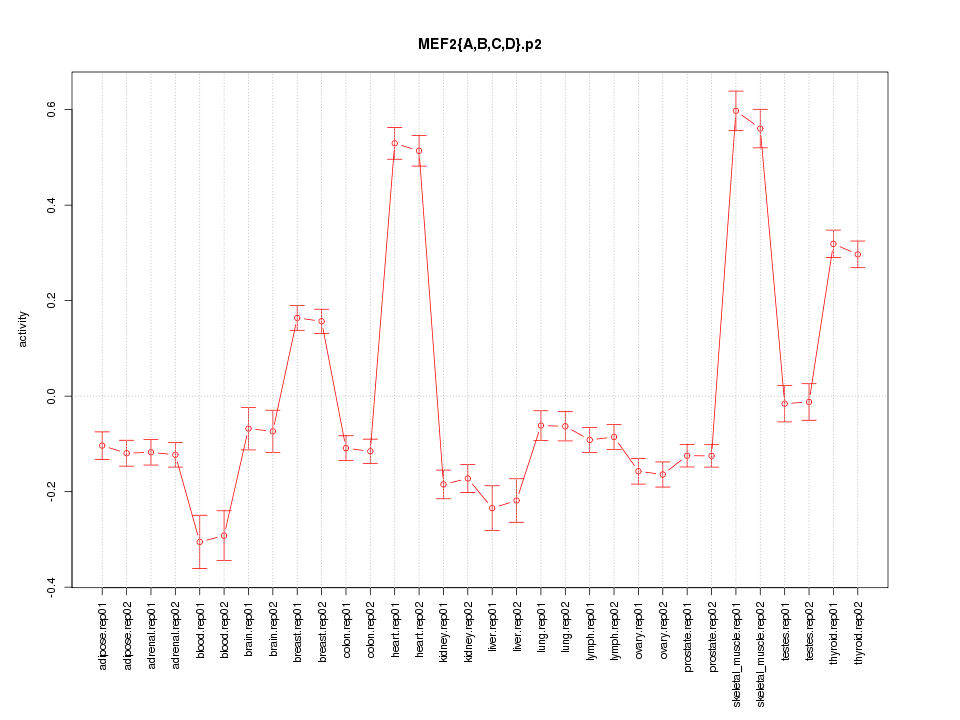
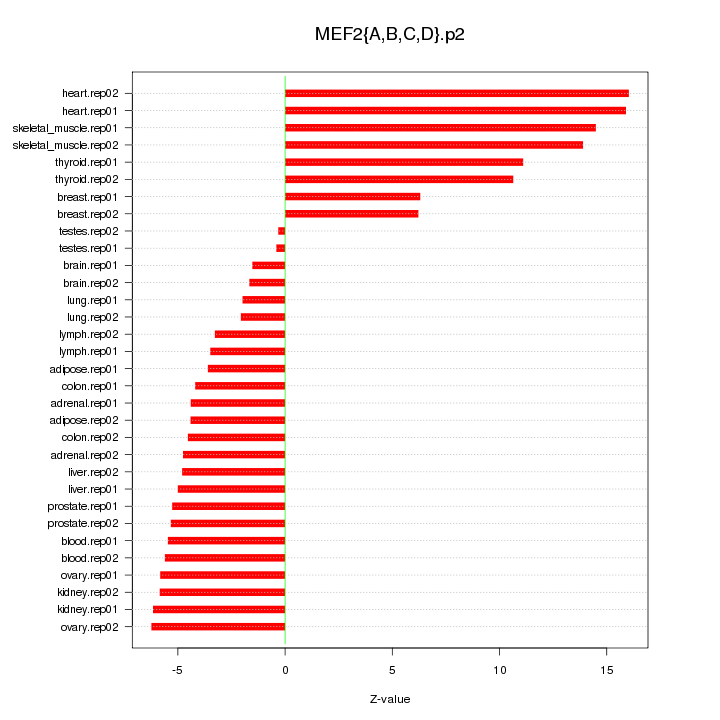
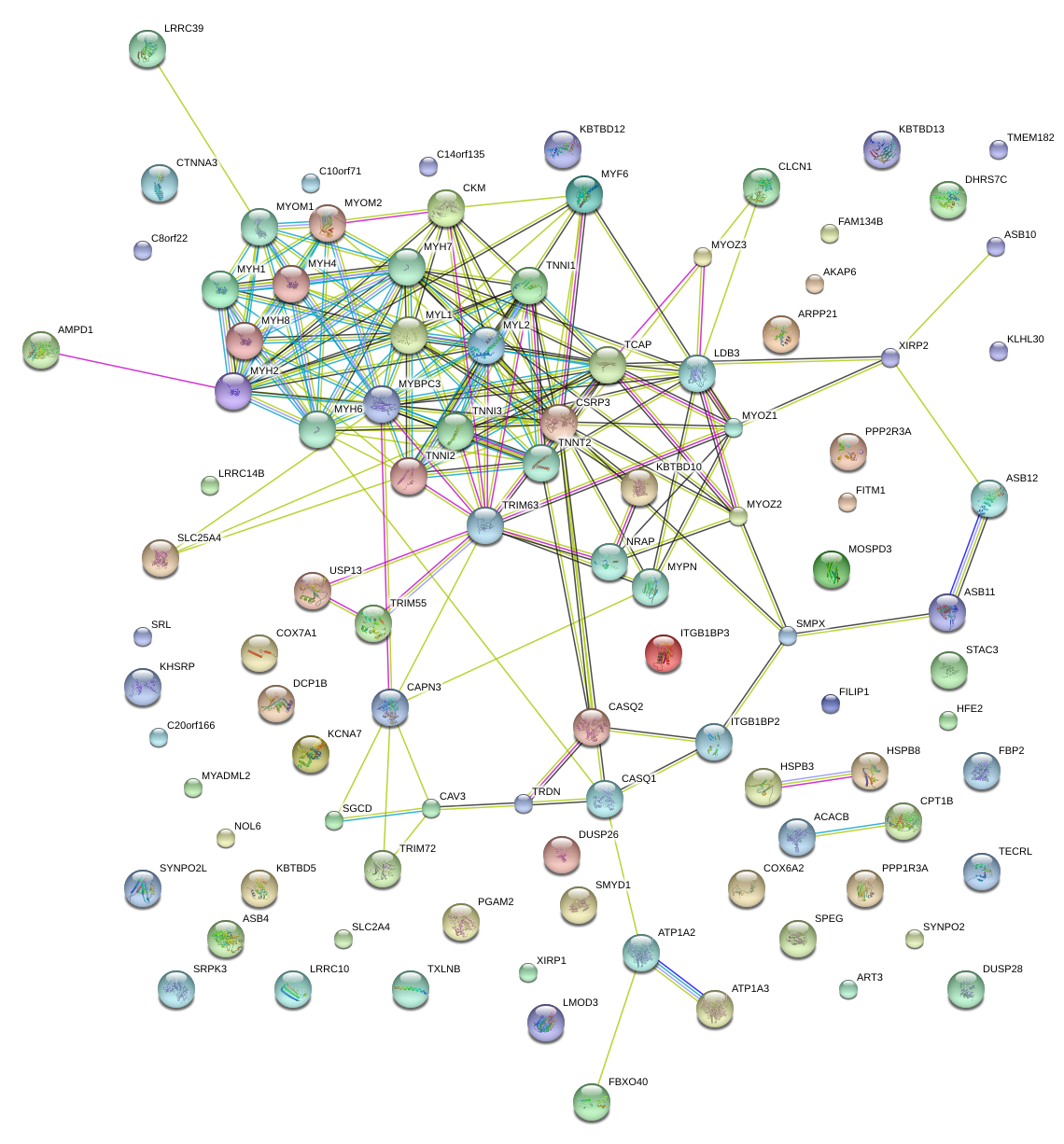
|
chr6_-_123957941
|
90.362
|
NM_001251987
NM_006073
|
TRDN
|
triadin
|
|
chr11_-_19223516
|
79.111
|
NM_001127656
NM_003476
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein)
|
|
chr7_-_113559048
|
76.510
|
NM_002711
|
PPP1R3A
|
protein phosphatase 1, regulatory subunit 3A
|
|
chr10_+_69869249
|
75.332
|
NM_032578
|
MYPN
|
myopalladin
|
|
chr17_+_37821598
|
71.056
|
NM_003673
|
TCAP
|
titin-cap (telethonin)
|
|
chr12_-_111358353
|
70.781
|
NM_000432
|
MYL2
|
myosin, light chain 2, regulatory, cardiac, slow
|
|
chrX_-_21776158
|
63.383
|
NM_014332
|
SMPX
|
small muscle protein, X-linked
|
|
chr2_+_88367298
|
59.452
|
NM_198274
|
SMYD1
|
SET and MYND domain containing 1
|
|
chr2_-_211168315
|
55.023
|
NM_079422
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr10_-_75415748
|
54.433
|
NM_001114133
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr3_+_121312169
|
53.214
|
NM_016298
|
FBXO40
|
F-box protein 40
|
|
chr3_-_39234071
|
53.201
|
NM_194293
NM_001198621
|
XIRP1
|
xin actin-binding repeat containing 1
|
|
chr2_+_168043792
|
50.502
|
NM_001199144
NM_001199145
|
XIRP2
|
xin actin-binding repeat containing 2
|
|
chr3_+_42727010
|
49.747
|
NM_152393
|
KBTBD5
|
kelch repeat and BTB (POZ) domain containing 5
|
|
chr14_-_23877480
|
49.419
|
NM_002471
|
MYH6
|
myosin, heavy chain 6, cardiac muscle, alpha
|
|
chr10_-_75410778
|
45.746
|
NM_024875
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr4_-_65275000
|
44.505
|
NM_001010874
|
TECRL
|
trans-2,3-enoyl-CoA reductase-like
|
|
chr12_+_81101407
|
43.527
|
NM_002469
|
MYF6
|
myogenic factor 6 (herculin)
|
|
chr18_-_3219986
|
41.664
|
NM_003803
NM_019856
|
MYOM1
|
myomesin 1, 185kDa
|
|
chr16_-_31439650
|
38.525
|
NM_005205
|
COX6A2
|
cytochrome c oxidase subunit VIa polypeptide 2
|
|
chr10_+_50507186
|
37.581
|
NM_001135196
NM_199459
|
C10orf71
|
chromosome 10 open reading frame 71
|
|
chr1_-_26393984
|
36.175
|
NM_032588
|
TRIM63
|
tripartite motif containing 63
|
|
chr18_-_3219846
|
35.159
|
|
MYOM1
|
myomesin 1, 185kDa
|
|
chr10_-_115423640
|
35.105
|
NM_006175
NM_198060
|
NRAP
|
nebulin-related anchoring protein
|
|
chr2_+_170366211
|
34.164
|
NM_006063
|
KBTBD10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr10_-_75401199
|
33.221
|
|
MYOZ1
|
myozenin 1
|
|
chr1_-_100643770
|
32.434
|
NM_144620
|
LRRC39
|
leucine rich repeat containing 39
|
|
chrX_-_15332680
|
32.138
|
NM_001012428
|
ASB11
|
ankyrin repeat and SOCS box containing 11
|
|
chr5_+_53751430
|
31.831
|
NM_006308
|
HSPB3
|
heat shock 27kDa protein 3
|
|
chr4_+_76995862
|
31.802
|
NM_001130016
NM_001179
|
ART3
|
ADP-ribosyltransferase 3
|
|
chr4_+_120056938
|
31.159
|
NM_016599
|
MYOZ2
|
myozenin 2
|
|
chr19_-_45822841
|
30.330
|
|
CKM
|
creatine kinase, muscle
|
|
chr7_+_95115212
|
30.318
|
NM_016116
NM_145872
|
ASB4
|
ankyrin repeat and SOCS box containing 4
|
|
chr1_-_201390845
|
29.480
|
NM_003281
|
TNNI1
|
troponin I type 1 (skeletal, slow)
|
|
chr1_+_145413190
|
29.433
|
NM_145277
NM_202004
NM_213652
NM_213653
|
HFE2
|
hemochromatosis type 2 (juvenile)
|
|
chr1_-_116311330
|
28.273
|
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr1_-_116311161
|
28.212
|
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr1_-_116311398
|
27.807
|
NM_001232
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr16_+_31225341
|
26.963
|
NM_001008274
|
TRIM72
|
tripartite motif containing 72
|
|
chr1_+_160160284
|
26.837
|
NM_001231
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle)
|
|
chr5_+_191625
|
26.578
|
NM_001080478
|
LRRC14B
|
leucine rich repeat containing 14B
|
|
chr17_-_10421858
|
25.962
|
NM_005963
|
MYH1
|
myosin, heavy chain 1, skeletal muscle, adult
|
|
chr16_-_4292052
|
25.803
|
NM_001098814
|
SRL
|
sarcalumenin
|
|
chr19_+_3933100
|
25.782
|
NM_170678
|
ITGB1BP3
|
integrin beta 1 binding protein 3
|
|
chr2_+_239047362
|
25.493
|
NM_198582
|
KLHL30
|
kelch-like 30 (Drosophila)
|
|
chr12_-_57644905
|
25.160
|
NM_145064
|
STAC3
|
SH3 and cysteine rich domain 3
|
|
chr10_-_69425356
|
24.704
|
NM_001127384
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr10_-_75401492
|
24.392
|
NM_021245
|
MYOZ1
|
myozenin 1
|
|
chr6_+_53883713
|
23.917
|
NM_138569
|
MLIP
|
muscular LMNA-interacting protein
|
|
chr15_+_65369153
|
23.291
|
NM_001101362
|
KBTBD13
|
kelch repeat and BTB (POZ) domain containing 13
|
|
chr17_-_9694613
|
23.265
|
NM_001105571
NM_001220493
|
DHRS7C
|
dehydrogenase/reductase (SDR family) member 7C
|
|
chr10_+_88428263
|
22.947
|
NM_001080116
NM_001080114
NM_001080115
NM_001171611
NM_007078
|
LDB3
|
LIM domain binding 3
|
|
chr17_+_7184978
|
22.789
|
NM_001042
|
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4
|
|
chr6_-_76203256
|
22.721
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr8_+_1993157
|
22.104
|
NM_003970
|
MYOM2
|
myomesin (M-protein) 2, 165kDa
|
|
chr1_+_160085519
|
21.456
|
NM_000702
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide
|
|
chr11_+_1860232
|
21.412
|
NM_003282
|
TNNI2
|
troponin I type 2 (skeletal, fast)
|
|
chr5_+_155753755
|
21.384
|
NM_000337
NM_001128209
NM_172244
|
SGCD
|
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein)
|
|
chr10_-_69455926
|
21.123
|
NM_013266
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr11_+_1860718
|
21.003
|
NM_001145829
|
TNNI2
|
troponin I type 2 (skeletal, fast)
|
|
chr2_+_103378489
|
20.117
|
NM_144632
|
TMEM182
|
transmembrane protein 182
|
|
chr8_-_33457491
|
19.711
|
|
DUSP26
|
dual specificity phosphatase 26 (putative)
|
|
chr3_+_8775485
|
19.555
|
NM_001234
NM_033337
|
CAV3
|
caveolin 3
|
|
chr3_+_35683848
|
19.371
|
NM_016300
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr7_-_150884477
|
19.218
|
NM_001142459
NM_001142460
|
ASB10
|
ankyrin repeat and SOCS box containing 10
|
|
chrX_-_63450486
|
19.169
|
NM_130388
|
ASB12
|
ankyrin repeat and SOCS box containing 12
|
|
chr5_-_16509106
|
19.061
|
NM_019000
|
FAM134B
|
family with sequence similarity 134, member B
|
|
chrX_+_153046422
|
18.965
|
NM_001170760
NM_001170761
NM_014370
|
SRPK3
|
SRSF protein kinase 3
|
|
chr1_-_115238091
|
18.689
|
NM_000036
NM_001172626
|
AMPD1
|
adenosine monophosphate deaminase 1
|
|
chr2_-_211179765
|
18.281
|
NM_079420
|
MYL1
|
myosin, light chain 1, alkali; skeletal, fast
|
|
chr7_-_150884918
|
17.927
|
NM_080871
|
ASB10
|
ankyrin repeat and SOCS box containing 10
|
|
chr17_-_79905108
|
17.923
|
NM_001145113
|
MYADML2
|
myeloid-associated differentiation marker-like 2
|
|
chr2_+_220299699
|
17.587
|
NM_005876
|
SPEG
|
SPEG complex locus
|
|
chr11_-_47374221
|
17.569
|
NM_000256
|
MYBPC3
|
myosin binding protein C, cardiac
|
|
chr3_+_135741576
|
17.502
|
NM_181897
|
PPP2R3A
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr20_+_61147659
|
17.340
|
NM_178463
|
C20orf166
|
chromosome 20 open reading frame 166
|
|
chr3_+_127634317
|
17.289
|
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chr19_-_49576120
|
17.245
|
NM_031886
|
KCNA7
|
potassium voltage-gated channel, shaker-related subfamily, member 7
|
|
chr6_-_139613114
|
17.059
|
NM_153235
|
TXLNB
|
taxilin beta
|
|
chr3_+_127641901
|
16.873
|
NM_207335
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chr14_+_24600674
|
16.494
|
NM_203402
|
FITM1
|
fat storage-inducing transmembrane protein 1
|
|
chr8_+_67039130
|
16.483
|
NM_033058
NM_184085
NM_184086
NM_184087
|
TRIM55
|
tripartite motif containing 55
|
|
chr19_-_55668956
|
16.408
|
NM_000363
|
TNNI3
|
troponin I type 3 (cardiac)
|
|
chr8_+_49984902
|
16.251
|
NM_001007176
|
C8orf22
|
chromosome 8 open reading frame 22
|
|
chr12_-_70004941
|
16.037
|
NM_201550
|
LRRC10
|
leucine rich repeat containing 10
|
|
chr22_-_51016845
|
15.882
|
NM_004377
NM_152246
NM_001145134
NM_001145135
NM_001145136
NM_152245
|
CPT1B
|
carnitine palmitoyltransferase 1B (muscle)
|
|
chr12_+_119616594
|
15.807
|
NM_014365
|
HSPB8
|
heat shock 22kDa protein 8
|
|
chr9_-_97356007
|
15.358
|
NM_003837
|
FBP2
|
fructose-1,6-bisphosphatase 2
|
|
chrX_+_70521627
|
15.171
|
NM_012278
|
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2
|
|
chr3_-_69171536
|
14.909
|
NM_198271
|
LMOD3
|
leiomodin 3 (fetal)
|
|
chr7_-_44105162
|
14.828
|
NM_000290
|
PGAM2
|
phosphoglycerate mutase 2 (muscle)
|
|
chr4_+_186064416
|
14.439
|
NM_001151
|
SLC25A4
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4
|
|
chr3_+_179370779
|
14.301
|
NM_003940
|
USP13
|
ubiquitin specific peptidase 13 (isopeptidase T-3)
|
|
chr5_+_150040402
|
14.248
|
NM_001122853
NM_133371
|
MYOZ3
|
myozenin 3
|
|
chr7_+_143013218
|
14.211
|
NM_000083
|
CLCN1
|
chloride channel 1, skeletal muscle
|
|
chr1_-_201346804
|
14.114
|
NM_000364
NM_001001430
NM_001001431
NM_001001432
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr19_-_36643770
|
14.052
|
NM_001864
|
COX7A1
|
cytochrome c oxidase subunit VIIa polypeptide 1 (muscle)
|
|
chr12_+_109554391
|
13.945
|
|
ACACB
|
acetyl-CoA carboxylase beta
|
|
chr14_+_32798478
|
13.909
|
NM_004274
|
AKAP6
|
A kinase (PRKA) anchor protein 6
|
|
chr4_+_119809978
|
13.816
|
NM_001128933
NM_001128934
NM_133477
|
SYNPO2
|
synaptopodin 2
|
|
chr8_-_33457279
|
13.751
|
|
DUSP26
|
dual specificity phosphatase 26 (putative)
|
|
chr15_+_42651697
|
13.683
|
NM_000070
NM_024344
NM_173087
|
CAPN3
|
calpain 3, (p94)
|
|
chr1_+_170633312
|
13.659
|
NM_006902
NM_022716
|
PRRX1
|
paired related homeobox 1
|
|
chr3_-_57326709
|
13.005
|
NM_001142733
|
ASB14
|
ankyrin repeat and SOCS box containing 14
|
|
chr8_-_107782417
|
12.993
|
NM_139166
|
ABRA
|
actin-binding Rho activating protein
|
|
chr4_+_110834039
|
12.956
|
NM_001178130
NM_001178131
NM_001963
|
EGF
|
epidermal growth factor
|
|
chr17_-_10372875
|
12.882
|
NM_017533
|
MYH4
|
myosin, heavy chain 4, skeletal muscle
|
|
chr7_+_123249076
|
12.854
|
NM_080928
|
ASB15
|
ankyrin repeat and SOCS box containing 15
|
|
chr1_-_201346783
|
12.833
|
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr14_-_94423766
|
12.222
|
NM_016150
|
ASB2
|
ankyrin repeat and SOCS box containing 2
|
|
chr4_+_95373044
|
12.051
|
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr8_-_82395401
|
11.872
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr7_+_28448970
|
11.758
|
|
LOC401317
|
uncharacterized LOC401317
|
|
chr12_-_22094166
|
11.612
|
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9
|
|
chr9_-_85882093
|
11.611
|
NM_001244962
|
FRMD3
|
FERM domain containing 3
|
|
chr10_+_88428167
|
11.572
|
NM_001171610
|
LDB3
|
LIM domain binding 3
|
|
chr2_+_233390869
|
11.548
|
NM_000751
|
CHRND
|
cholinergic receptor, nicotinic, delta
|
|
chr17_+_45286713
|
11.382
|
NM_002476
|
MYL4
|
myosin, light chain 4, alkali; atrial, embryonic
|
|
chr3_-_196242190
|
11.300
|
NM_001077657
|
C3orf43
|
chromosome 3 open reading frame 43
|
|
chr3_-_192445344
|
11.169
|
NM_004113
|
FGF12
|
fibroblast growth factor 12
|
|
chr5_+_137203544
|
11.140
|
NM_001135940
NM_006790
|
MYOT
|
myotilin
|
|
chr12_+_109665175
|
11.088
|
|
ACACB
|
acetyl-CoA carboxylase beta
|
|
chr11_-_101787252
|
11.015
|
NM_178127
|
ANGPTL5
|
angiopoietin-like 5
|
|
chr12_-_70093064
|
10.979
|
NM_032735
|
BEST3
|
bestrophin 3
|
|
chr4_-_177190258
|
10.819
|
NM_080874
|
ASB5
|
ankyrin repeat and SOCS box containing 5
|
|
chr17_-_10325266
|
10.640
|
NM_002472
|
MYH8
|
myosin, heavy chain 8, skeletal muscle, perinatal
|
|
chr17_+_42248073
|
10.606
|
NM_080863
|
ASB16
|
ankyrin repeat and SOCS box containing 16
|
|
chr19_-_49575878
|
10.599
|
|
KCNA7
|
potassium voltage-gated channel, shaker-related subfamily, member 7
|
|
chr4_+_41362843
|
10.523
|
|
|
|
|
chr5_+_149997205
|
10.367
|
NM_001166209
|
SYNPO
|
synaptopodin
|
|
chr10_-_92681025
|
10.177
|
NM_014391
|
ANKRD1
|
ankyrin repeat domain 1 (cardiac muscle)
|
|
chr9_-_130639939
|
9.851
|
NM_000476
|
AK1
|
adenylate kinase 1
|
|
chr4_+_41362565
|
9.848
|
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr4_+_41362750
|
9.836
|
NM_001112717
NM_001112718
NM_014988
|
LIMCH1
|
LIM and calponin homology domains 1
|
|
chr2_-_179914785
|
9.759
|
NM_173648
|
CCDC141
|
coiled-coil domain containing 141
|
|
chr3_+_35721115
|
9.678
|
NM_001025068
NM_001025069
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr15_+_78558522
|
9.651
|
NM_001130183
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr4_-_23891608
|
9.463
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr1_-_31845818
|
9.384
|
NM_004102
|
FABP3
|
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor)
|
|
chr18_-_24445652
|
9.205
|
NM_001650
|
AQP4
|
aquaporin 4
|
|
chr17_+_43971655
|
9.169
|
NM_001123066
NM_001123067
NM_001203251
NM_001203252
NM_005910
NM_016834
NM_016835
NM_016841
|
MAPT
|
microtubule-associated protein tau
|
|
chr5_-_142077508
|
9.140
|
NM_001144934
NM_001144935
|
FGF1
|
fibroblast growth factor 1 (acidic)
|
|
chr16_+_28891234
|
8.932
|
|
ATP2A1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1
|
|
chr8_-_70745624
|
8.925
|
NM_001146008
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr15_-_70994608
|
8.886
|
NM_001008224
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr8_-_27850368
|
8.864
|
NM_173833
|
SCARA5
|
scavenger receptor class A, member 5 (putative)
|
|
chr7_-_15726026
|
8.606
|
|
MEOX2
|
mesenchyme homeobox 2
|
|
chr3_-_195310770
|
8.591
|
|
APOD
|
apolipoprotein D
|
|
chr1_-_205912546
|
8.452
|
NM_052934
NM_134325
|
SLC26A9
|
solute carrier family 26, member 9
|
|
chr4_+_95373007
|
8.423
|
NM_001011513
NM_001011515
NM_001011516
NM_006457
|
PDLIM5
|
PDZ and LIM domain 5
|
|
chr12_-_49392909
|
8.395
|
NM_015086
|
DDN
|
dendrin
|
|
chr13_+_76334796
|
8.354
|
NM_015842
|
LMO7
|
LIM domain 7
|
|
chrX_-_15333726
|
8.313
|
NM_001201583
NM_080873
|
ASB11
|
ankyrin repeat and SOCS box containing 11
|
|
chr8_-_72268720
|
8.312
|
NM_172058
NM_172059
NM_172060
|
EYA1
|
eyes absent homolog 1 (Drosophila)
|
|
chr7_-_15726273
|
8.266
|
NM_005924
|
MEOX2
|
mesenchyme homeobox 2
|
|
chr1_-_217113007
|
8.212
|
NM_001243512
NM_001243513
NM_001243510
NM_001243511
|
ESRRG
|
estrogen-related receptor gamma
|
|
chr9_+_100263818
|
8.082
|
NM_003275
|
TMOD1
|
tropomodulin 1
|
|
chr16_+_30075440
|
7.734
|
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr16_+_30075530
|
7.731
|
NM_184043
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr8_-_23712297
|
7.674
|
NM_003155
|
STC1
|
stanniocalcin 1
|
|
chr14_-_70546863
|
7.639
|
NM_182936
NM_001130417
|
SLC8A3
|
solute carrier family 8 (sodium/calcium exchanger), member 3
|
|
chr6_-_2751153
|
7.546
|
NM_001012418
|
MYLK4
|
myosin light chain kinase family, member 4
|
|
chr2_-_106055229
|
7.514
|
NM_201557
|
FHL2
|
four and a half LIM domains 2
|
|
chr11_+_3666357
|
7.491
|
NM_004314
|
ART1
|
ADP-ribosyltransferase 1
|
|
chr4_-_186697065
|
7.471
|
NM_001145673
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr3_-_47933997
|
7.441
|
|
MAP4
|
microtubule-associated protein 4
|
|
chr20_+_44035203
|
7.440
|
NM_001048223
NM_001048224
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2
|
|
chr17_-_67057046
|
7.428
|
NM_080283
|
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9
|
|
chr7_-_30739660
|
7.321
|
NM_001202475
NM_001202481
|
CRHR2
|
corticotropin releasing hormone receptor 2
|
|
chr1_+_13911966
|
7.299
|
NM_001006624
NM_001006625
|
PDPN
|
podoplanin
|
|
chr1_-_23751126
|
7.207
|
NM_003196
|
TCEA3
|
transcription elongation factor A (SII), 3
|
|
chr5_+_149980627
|
7.094
|
NM_001166208
|
SYNPO
|
synaptopodin
|
|
chr3_+_189507448
|
7.085
|
NM_001114980
NM_001114981
NM_001114982
|
TP63
|
tumor protein p63
|
|
chr3_-_100712248
|
7.066
|
NM_015429
|
ABI3BP
|
ABI family, member 3 (NESH) binding protein
|
|
chr11_-_62457001
|
7.058
|
|
LRRN4CL
|
LRRN4 C-terminal like
|
|
chr19_-_48614013
|
6.959
|
NM_001159322
NM_001159323
NM_003706
|
PLA2G4C
|
phospholipase A2, group IVC (cytosolic, calcium-independent)
|
|
chr1_+_74701070
|
6.921
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr3_+_69812961
|
6.909
|
NM_006722
|
MITF
|
microphthalmia-associated transcription factor
|
|
chr1_+_15990300
|
6.905
|
|
|
|
|
chr7_+_80231503
|
6.868
|
NM_001001547
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr9_+_124048396
|
6.846
|
|
GSN
|
gelsolin
|
|
chr8_-_62602285
|
6.827
|
NM_001164750
NM_001164751
NM_001164752
NM_001164753
NM_020164
NM_032467
NM_032468
|
ASPH
|
aspartate beta-hydroxylase
|
|
chr1_-_16344438
|
6.773
|
|
HSPB7
|
heat shock 27kDa protein family, member 7 (cardiovascular)
|
|
chr1_-_24438625
|
6.752
|
NM_152372
|
MYOM3
|
myomesin family, member 3
|
|
chr4_+_113970784
|
6.748
|
NM_001148
NM_020977
|
ANK2
|
ankyrin 2, neuronal
|
|
chr4_+_169418167
|
6.724
|
NM_001166108
NM_016081
|
PALLD
|
palladin, cytoskeletal associated protein
|
|
chr3_+_99357439
|
6.667
|
NM_001850
NM_020351
|
COL8A1
|
collagen, type VIII, alpha 1
|
|
chr14_-_105634708
|
6.646
|
|
JAG2
|
jagged 2
|
|
chr3_+_50316517
|
6.596
|
NM_153215
|
C3orf45
|
chromosome 3 open reading frame 45
|
|
chr2_-_217560247
|
6.580
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr10_+_86184685
|
6.572
|
|
FAM190B
|
family with sequence similarity 190, member B
|
|
chr2_-_237172987
|
6.569
|
NM_212556
|
ASB18
|
ankyrin repeat and SOCS box containing 18
|
|
chr16_+_30075715
|
6.544
|
NM_001127617
|
ALDOA
|
aldolase A, fructose-bisphosphate
|
|
chr7_+_75931991
|
6.526
|
|
HSPB1
|
heat shock 27kDa protein 1
|
|
chr2_-_152590983
|
6.459
|
NM_001164507
NM_001164508
NM_004543
|
NEB
|
nebulin
|
|
chr2_+_168675181
|
6.433
|
NM_020981
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1
|
|
chr3_+_99833773
|
6.376
|
NM_001167924
|
C3orf26
|
chromosome 3 open reading frame 26
|
|
chr6_+_12716887
|
6.332
|
|
PHACTR1
|
phosphatase and actin regulator 1
|
|
chr20_+_9288446
|
6.179
|
NM_000933
|
PLCB4
|
phospholipase C, beta 4
|
|
chr3_-_152058778
|
6.131
|
NM_001123228
|
TMEM14E
|
transmembrane protein 14E
|